Wednesday, December 29, 2010
install srma
Friday, December 10, 2010
Common indicators of variant quality
7550 Benchmark
Thursday, December 9, 2010
HapMap
The DNA in our cells contains long chains of four chemical building blocks -- adenine, thymine, cytosine, and guanine, abbreviated A, T, C, and G. More than 6 billion of these chemical bases, strung together in 23 pairs of chromosomes, exist in a human cell. (See http://www.dnaftb.org/dnaftb/ for basic information about genetics.) These genetic sequences contain information that influences our physical traits, our likelihood of suffering from disease, and the responses of our bodies to substances that we encounter in the environment.
The genetic sequences of different people are remarkably similar. When the chromosomes of two humans are compared, their DNA sequences can be identical for hundreds of bases. But at about one in every 1,200 bases, on average, the sequences will differ (Figure 1). One person might have an A at that location, while another person has a G, or a person might have extra bases at a given location or a missing segment of DNA. Each distinct "spelling" of a chromosomal region is called an allele, and a collection of alleles in a person's chromosomes is known as a genotype.
Differences in individual bases are by far the most common type of genetic variation. These genetic differences are known as single nucleotide polymorphisms, or SNPs (pronounced "snips"). By identifying most of the approximately 10 million SNPs estimated to occur commonly in the human genome, the International HapMap Project is identifying the basis for a large fraction of the genetic diversity in the human species.
For geneticists, SNPs act as markers to locate genes in DNA sequences. Say that a spelling change in a gene increases the risk of suffering from high blood pressure, but researchers do not know where in our chromosomes that gene is located. They could compare the SNPs in people who have high blood pressure with the SNPs of people who do not. If a particular SNP is more common among people with hypertension, that SNP could be used as a pointer to locate and identify the gene involved in the disease.
However, testing all of the 10 million common SNPs in a person's chromosomes would be extremely expensive. The development of the HapMap will enable geneticists to take advantage of how SNPs and other genetic variants are organized on chromosomes. Genetic variants that are near each other tend to be inherited together. For example, all of the people who have an A rather than a G at a particular location in a chromosome can have identical genetic variants at other SNPs in the chromosomal region surrounding the A. These regions of linked variants are known as haplotypes.
In many parts of our chromosomes, just a handful of haplotypes are found in humans. [See The Origins of Haplotypes.] In a given population, 55 percent of people may have one version of a haplotype, 30 percent may have another, 8 percent may have a third, and the rest may have a variety of less common haplotypes. The International HapMap Project is identifying these common haplotypes in four populations from different parts of the world. It also is identifying "tag" SNPs that uniquely identify these haplotypes. By testing an individual's tag SNPs (a process known as genotyping), researchers will be able to identify the collection of haplotypes in a person's DNA. The number of tag SNPs that contain most of the information about the patterns of genetic variation is estimated to be about 300,000 to 600,000, which is far fewer than the 10 million common SNPs.
Once the information on tag SNPs from the HapMap is available, researchers will be able to use them to locate genes involved in medically important traits. Consider the researcher trying to find genetic variants associated with high blood pressure. Instead of determining the identity of all SNPs in a person's DNA, the researcher would genotype a much smaller number of tag SNPs to determine the collection of haplotypes present in each subject. The researcher could focus on specific candidate genes that may be associated with a disease, or even look across the entire genome to find chromosomal regions that may be associated with a disease. If people with high blood pressure tend to share a particular haplotype, variants contributing to the disease might be somewhere within or near that haplotype.
Transition and transcersion
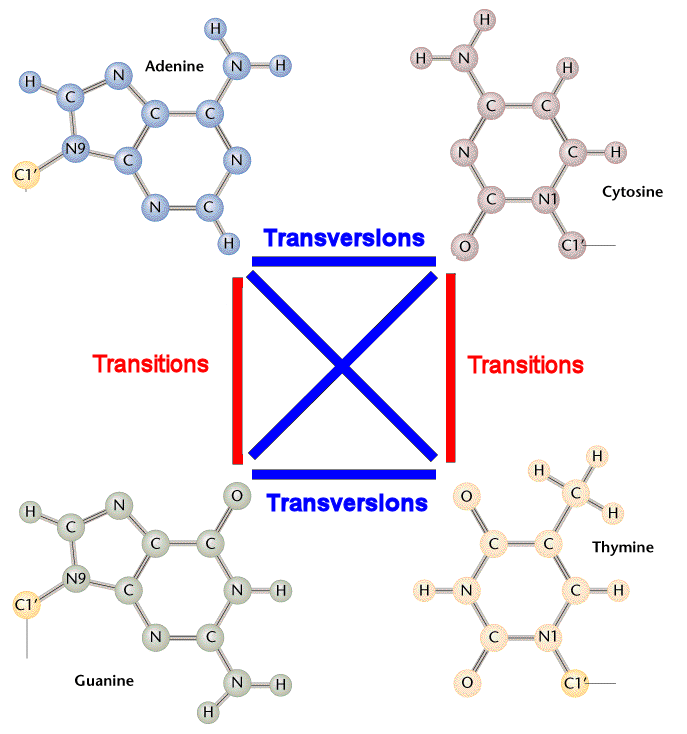
DNA substitution mutations are of two types. Transitions are interchanges of two-ring purines (A G) or of one-ring pyrimidines (C T): they therefore involve bases of similar shape. Transversions are interchanges of purine for pyrimidine bases, which therefore involve exchange of one-ring and two-ring structures.
Although there are twice as many possible transversions, because of the molecular mechanisms by which they are generated, transition mutations are generated at higher frequency than transversions. As well, transitions are less likely to result in amino acid substitutions (due to "wobble"), and are therefore more likely to persist as "silent substitutions" in populations as single nucleotide polymorphisms (SNPs).
Wednesday, December 8, 2010
make a simulation for 7550R
Monday, December 6, 2010
clustered SNP
Thursday, December 2, 2010
Read group
For example, to create a paired end same file with fully formatted read groups, you can run:
bwa sampe -i MARK -m DEPRISTO -l LIBRARY -p ILLUMINA -h Homo_sapiens_assembly18.fasta first.fastq.sai secondfastq.sai first.fastq second.fastq > pe.sam
This will result in a file that looks like:
@HD VN:1.0 SO:unsorted
@SQ SN:chrM LN:16571
@SQ SN:chr1 LN:247249719
@SQ SN:chr2 LN:242951149
[More @SQ lines removed for clarity]
@RG ID:MARK SM:DEPRISTO LB:LIBRARY PL:ILLUMINA
61CC3AAXX100125:6:48:11631:21212 81 chrM 8 37 76M chr17 21944837 0 GGTCTATCACCCTATTAACCACTCACGGGAGCTCTCCATGCATTTGGTATTTTCGTCTGGGGGGTATGCACGCGAT 99:<->=D>IF7.:7,H
Tuesday, November 30, 2010
dbSNP file naming convention
Friday, November 12, 2010
What is base calling?
Monday, November 8, 2010
Pileup format
SNP call
Tuesday, November 2, 2010
delete lines from a file
soft clipped
Monday, October 18, 2010
A SNP Call pipeline
SNP and MicroIndel detection
Novoalign detects mismatches and short indels in short reads from next-generation sequencing platforms. The recipes below contain command line usage to generate list of single nucleotide polymorphisms SNPs and Indels for further analysis.
Generate sorted alignments in BAM format
Inititally we need to align our file(s) to the reference genome:
novoalign -d hg18.nix -f reads1.fastq -o SAM 2> stats.txt > novo.sam
samtools view -bS novo.sam | samtools sort - reads1
For paired-end or mate-pair reads adjust the parameters accordingly. If we have multiple alignment files these all need to be sorted individually and merged with either samtools or Picard.
Remove PCR duplicates
It is highly recommended to remove PCR duplicates before proceeding with the SNP and Indel calls. We use samtools as an example to remove PCR duplicates.
#remove PCR duplicates from single-end reads
samtools rmdup -s reads1.bam reads1.sorted.bam
#For Paired reads
samtools rmdup reads1.bam reads1.pe.sorted.bam
SNP and Microindel calling
In this example we use the samtools pileup SNP caller and accept reads with a minimum mapping quality of 20:
#The hg18.fasta reference genome sequence is required. samtools.pl is also required
samtools view -u -q 20 reads1.bam | samtools pileup -vc -f hg18.fasta - | samtools.pl varFilter -D 1000 -G 50 > variations.list
Filtering and final list generation
A simple awk script can be used to filter our list of variations where the indel SNP quality is greater than 50 or 60 respectively:
awk '($3=="*"&&$6>=50)||($3!="*"&&$6>30)' variations.list > variations.filtered.list
Exporting to other formats
The samtools distribution contains a helper script called sam2vcf.pl that may be used to convert to the Variant Call Format (VCF):
sam2vcf.pl variations.filtered.list -r hg18.fasta > variations.vcf
Thursday, October 7, 2010
Walltime estimation
Tuesday, September 21, 2010
Simulate data for pipeline
Wednesday, September 15, 2010
update potato source code
Tuesday, September 14, 2010
delete all jobs with name=hello
Friday, September 10, 2010
Supported Genome Builds
Human (hg19, Feb_2009, GRCh37)
single-end, HiSeq2000, Rong Mao Lab
Human (hg18, Mar_2006, NCBI Build 36.1)
DNA-methylation, pair-end, Hiseq2000, Brad Cairns Lab
Chip-seq, single-end, GAIIx, Brad Cairns Lab
C_elegans (CE6, May_2008, WS190)
Small RNA sequencing, single-end, GAIIx, Brenda Bass Lab
A_Thaliana (TAIR8, Mar_2008)
Single-end, HiSeq2000, Jason Stajich Lab
M_musculus (mm9, Jul_2007, NCBI Build 37)
ChIP-Seq, single-end, HiSeq2000, Anne Moon Lab
DNA-methylation, pair-end, HiSeq2000, Brad Cairns Lab
D_rerio (Zv8, Jun_2008)
ChIP-Seq, single-end, HiSeq2000, Brad Cairns Lab
Friday, September 3, 2010
No available computational resource at all
Torque and S3cmd
Wednesday, September 1, 2010
Amazon Cloud
Tuesday, August 31, 2010
Friday, August 27, 2010
Tuesday, August 24, 2010
Script mapping
novoalign -F ILMFQ -t120 -r0.2 -q5 -d hg19Splices34.index -f 1.txt 2.txt > 84060.novoalign
#transform to MAQ
maq novo2maq 84060.novoalign.map - 84060.novoalign
#Convert the reference sequences to the binary fasta format
maq fasta2bfa ref.fasta ref.bfa
#Build the mapping assembly
maq assemble consensus.cns ref.bfa 84060.novoalign.map 2>assemble.log
#Extract consensus sequences and qualities
maq cns2fq consensus.cns >cns.fq
#Extract list of SNPs
maq cns2snp consensus.cns >cns.snp
will be translated to :
/PATH_TO_NONOALIGN/novoalign -F ILMFQ -t120 -r0.2 -q5 -d /PATH_TO_GENOME_INDEX/hg19Splices34.index -f /PATH_TO_INPUT_FILE/1.txt /PATH_TO_INPUT_FILE/2.txt > /PATH_TO_OUTPUT_FILE/1_84060.novoalign
/PATH_TO_MAQ/maq novo2maq /PATH_TO_OUTPUT_FILE/84060.novoalign.map - /PATH_TO_INPUT_FILE/84060.novoalign
...
Monday, August 23, 2010
query, download, upload and update with rsync
#!/home/ying/py2.6.5/bin/python
"""
Use rsync to synchronize the DATA_HOME(hci-bio2, which raw data was stored) and COMP_HOME(chpc-node,
which process raw data and return output back to source. All the rsync operations are initiated from chpc-node.
Functions:
1. query: query the source if the new data is available
2. download: download new data ( from source to destination)
3. upload: upload result(output) to source.
4. update: update the source by appending a log file from the destination to show the alignment progress on chpc. "begin, percent-finished, end, error, etc"
"""
import os
import time
import operator
import subprocess
DATA_HOME = 'ying@123.123.123.123:~/alignment/'
COMP_HOME = '/home/ying/alignment/'
RSYNC_QUERY_PARAMS = '-are ssh'
RYSNC_DOWNLOAD_PARAMS = '-avuze ssh' #archive, verbose, update-only, compress-during-transfer and trasnfer-over-ssh
RYSNC_UPLOAD_PARAMS = '-avuze ssh' #archive, verbose, update-only, compress-during-transfer and trasnfer-over-ssh
LOG_FILE_NAME = 'log.txt'
def query():
#query_cmd = """rsync -are ssh ying@155.100.235.73:~/alignment/ | awk '{print $1 " " $3"#"$4 " " $5}'"""
cmd = ["rsync",RSYNC_QUERY_PARAMS,DATA_HOME,'|',"""awk '{print $1 " " $3"#"$4 " " $5}'"""]
query_cmd = ' '.join(cmd)
#print query_cmd
p = subprocess.Popen(query_cmd,shell=True,stdout=subprocess.PIPE)
line = p.communicate()
fnames = ''.join(line[:-1]).strip().split('\n')
df = {}
dt = {}
for fname in fnames:
prop,ctime,fn = fname.split(' ')
if fn != '.' and fn != '..':
if prop[0] == 'd': #it is a directory
pass
if prop[0] == '-': #it is a file
parent_path = os.path.dirname(fn)
df.setdefault(parent_path,[]).append(os.path.basename(fn))
#Transform time to seconds, for comparing which job should be run firstly. First In First Run.
#'2010/08/20#16:27:40' -> 1282343260.0
dt[parent_path] = int(time.mktime(time.strptime(ctime,'%Y/%m/%d#%H:%M:%S')))
jobs = []
for k,v in df.items():
if "begin" in v: #the data is ready
if not LOG_FILE_NAME in v: #this job is not running
jobs.append((k,dt[k]))
#print jobs
return sorted(jobs,key=operator.itemgetter(1))
def download(path_to_download):
"""download a whole directory from DATA_HOME to COMP_HOME
"""
normalized_path = path_to_download.strip('/') #/A, /A/ or A/ -> A
source = os.path.join(DATA_HOME,normalized_path)
cmd = ["rsync", RYSNC_DOWNLOAD_PARAMS, source, COMP_HOME] #download the whole directory to COMP_HOME.
download_cmd = ' '.join(cmd)
#print download_cmd
p = subprocess.Popen(download_cmd,shell=True,stdout=subprocess.PIPE)
line = p.communicate()
stdout_message = ''.join(line[:-1]).strip().split('\n')
#print 'Message:',stdout_message
update(normalized_path,stdout_message)
def upload(path,file_to_upload):
"""upload a file from COMP_HOME to DATA_HOME
upload 'file_to_upload' at local path 'folder'.
:/folder/file_to_upload will be uploaded to :/
"""
normalized_path = path.strip('/')+"/" #/A, /A/ or A/ -> A/
source = os.path.join(COMP_HOME,normalized_path,file_to_upload)
dest = os.path.join(DATA_HOME,normalized_path)
cmd = ["rsync",RYSNC_UPLOAD_PARAMS, source,dest]
upload_cmd = ' '.join(cmd)
p = subprocess.Popen(upload_cmd,shell=True,stdout=subprocess.PIPE)
line = p.communicate()
stdout_message = ''.join(line[:-1]).strip().split('\n')
#fn_log = log(os.path.join(COMP_HOME,normalized_src),stdout_message)
update(normalized_path,stdout_message)
def update(path,message):
fn = os.path.join(COMP_HOME,path,LOG_FILE_NAME)
ofn = None
try:
if os.path.exists(fn):
ofn = open(fn,'w+')#append
else:
ofn = open(fn,'w') #new file
print >>ofn,'-'*50
print >>ofn,time.asctime()
for m in message:
print >>ofn,M
ofn.close()
normalized_path = path.strip('/')+"/" #/A, /A/ or A/ -> A/
source = fn
dest = os.path.join(DATA_HOME,normalized_path)
cmd = ["rsync",RYSNC_UPLOAD_PARAMS, source,dest]
subprocess.Popen(cmd,shell=False,stdout=subprocess.PIPE)
except:
pass
def clear():
pass
#print query()
#download('A')
upload('A','hello.txt')